-Search query
-Search result
Showing all 30 items for (author: quemin & erj)

EMDB-18963: 
Structure of the SFTSV L protein in a transcription-priming state without capped RNA [TRANSCRIPTION-PRIMING (in vitro)]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Busch C, Milewski M, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-18967: 
Structure of the SFTSV L protein in a transcription-priming state with bound capped RNA [TRANSCRIPTION-PRIMING]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Busch C, Milewski M, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-18969: 
Structure of the SFTSV L protein stalled in a transcription-specific early elongation state with bound capped RNA [TRANSCRIPTION-EARLY-ELONGATION]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Busch C, Milewski M, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M
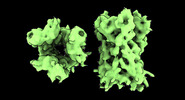
EMDB-17704: 
Subtomogram average of Vaccinia A10 trimer with open center from in vitro cores
Method: subtomogram averaging / : Turonova B, Liu J
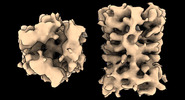
EMDB-17708: 
Subtomogram average of Vaccinia A10 trimer with tight center from in vitro cores
Method: subtomogram averaging / : Turonova B, Liu J

EMDB-17753: 
Subtomogram average of Vaccinia A10 trimer from in situ cores
Method: subtomogram averaging / : Turonova B, Liu J

EMDB-16670: 
Structure of the SNV L protein bound to 5' RNA
Method: single particle / : Meier K, Thorkelsson SR, Durieux Trouilleton Q, Vogel D, Yu D, Kosinski J, Cusack S, Malet H, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15607: 
Structure of the SFTSV L protein bound to 5' cRNA hook [5' HOOK]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15608: 
Structure of the SFTSV L protein stalled at early elongation [EARLY-ELONGATION]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15610: 
Structure of the SFTSV L protein stalled at early elongation with the endonuclease domain in a raised conformation [EARLY-ELONGATION-ENDO]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15614: 
Structure of the SFTSV L protein stalled at late elongation [LATE-ELONGATION]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15615: 
Structure of the SFTSV L protein bound in a resting state [RESTING]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15627: 
Tomogram of the Mimivirus genomic fiber
Method: electron tomography / : Villalta A, Schmitt A, Estrozi L, Quemin ERK, Alempic JM, Lartigue A, Prazak V, Belmudes L, Vasishtan D, Colmant AMG, Honore FA, Coute Y, Gruenewald K, Abergel C

EMDB-15628: 
Tomogram of the Mimivirus genomic fiber
Method: electron tomography / : Villalta A, Schmitt A, Estrozi L, Quemin ERK, Alempic JM, Lartigue A, Prazak V, Belmudes L, Vasishtan D, Colmant AMG, Honore FA, Coute Y, Gruenewald K, Abergel C

EMDB-15629: 
Tomogram of the Mimivirus genomic fiber
Method: electron tomography / : Villalta A, Schmitt A, Estrozi L, Quemin ERK, Alempic JM, Lartigue A, Prazak V, Belmudes L, Vasishtan D, Colmant AMG, Honore FA, Coute Y, Gruenewald K, Abergel C

EMDB-15630: 
Tomogram of the Mimivirus genomic fiber
Method: electron tomography / : Villalta A, Schmitt A, Estrozi L, Quemin ERK, Alempic JM, Lartigue A, Prazak V, Belmudes L, Vasishtan D, Colmant AMG, Honore FA, Coute Y, Gruenewald K, Abergel C

EMDB-13641: 
Structure of the Mimivirus genomic fibre asymmetric unit
Method: single particle / : Villalta A, Schmitt A, Estrozi LF, Quemin ERJ, Alempic JM, Lartigue A, Prazak V, Belmudes L, Vasishtan D, Colmant AMG, Honore FA, Coute Y, Grunewald K, Abergel C

EMDB-14353: 
Structure of the Mimivirus genomic fibre in its compact 6-start helix form
Method: helical / : Villalta A, Schmitt A

EMDB-14354: 
Structure of the Mimivirus genomic fibre in its compact 5-start helix form
Method: helical / : Villalta A, Schmitt A, Estrozi LF, Quemin ERJ, Alempic JM, Lartigue A, Prazak V, Belmudes L, Vasishtan D, Colmant AMG, Honore FA, Coute Y, Grunewald K, Abergel C

EMDB-14355: 
Structure of the Mimivirus genomic fibre in its relaxed 5-start helix form
Method: helical / : Villalta A, Schmitt A, Estrozi LF, Quemin ERJ, Alempic JM, Lartigue A, Prazak V, Belmudes L, Vasishtan D, Colmant AMG, Honore FA, Coute Y, Grunewald K, Abergel C

EMDB-12807: 
Apo-structure of Lassa virus L protein (well-resolved polymerase core) [APO-CORE]
Method: single particle / : Kouba T, Vogel D, Thorkelsson S, Quemin E, William H, Milewski M, Busch C, Gunther S, Grunewald K, Rosenthal M, Cusack S

EMDB-12860: 
Apo-structure of Lassa virus L protein (well-resolved endonuclease) [APO-ENDO]
Method: single particle / : Kouba T, Vogel D, Thorkelsson S, Quemin E, William H, Milewski M, Busch C, Gunther S, Grunewald K, Rosenthal M, Cusack S

EMDB-12861: 
Apo-structure of Lassa virus L protein (well-resolved alpha ribbon) [APO-RIBBON]
Method: single particle / : Kouba T, Vogel D, Thorkelsson S, Quemin E, Williams H, Milewski M, Busch C, Gunther S, Grunewald K, Rosenthal M, Cusack S

EMDB-12862: 
Lassa virus L protein bound to 3' promoter RNA (well-resolved polymerase core) [3END-CORE]
Method: single particle / : Kouba T, Vogel D, Thorkelsson S, Quemin E, William H, Milewski M, Busch C, Gunther S, Grunewald K, Rosenthal M, Cusack S

EMDB-12863: 
Lassa virus L protein bound to 3' promoter RNA (well-resolved endonuclease) [3END-ENDO]
Method: single particle / : Kouba T, Vogel D, Thorkelsson S, Quemin E, William H, Milewski M, Busch C, Gunther S, Grunewald K, Rosenthal M, Cusack S

EMDB-12953: 
Lassa virus L protein with endonuclease and C-terminal domains in close proximity [MID-LINK]
Method: single particle / : Kouba T, Vogel D, Thorkelsson S, Quemin E, William H, Milewski M, Busch C, Gunther S, Grunewald K, Rosenthal M, Cusack S

EMDB-12954: 
Lassa virus L protein bound to the distal promoter duplex [DISTAL-PROMOTER]
Method: single particle / : Kouba T, Vogel D, Thorkelsson S, Quemin E, William H, Milewski M, Busch C, Gunther S, Grunewald K, Rosenthal M, Cusack S

EMDB-12955: 
Lassa virus L protein in a pre-initiation conformation [PREINITIATION]
Method: single particle / : Kouba T, Vogel D, Thorkelsson S, Quemin E, William H, Milewski M, Busch C, Gunther S, Grunewald K, Rosenthal M, Cusack S

EMDB-12956: 
Lassa virus L protein in an elongation conformation [ELONGATION]
Method: single particle / : Kouba T, Vogel D, Thorkelsson S, Quemin E, William H, Milewski M, Busch C, Gunther S, Grunewald K, Rosenthal M, Cusack S

EMDB-10706: 
Cryo-EM structure of a Phenuiviridae L protein
Method: single particle / : Vogel D, Thorkelsson SR
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
